WELCOME TO OUR BLOG

Biomage is Acquired by Parse Biosciences
We are excited to announce that Biomage has been acquired by Parse Biosciences, the leading provider of accessible and scalable single cell sequencing solutions!
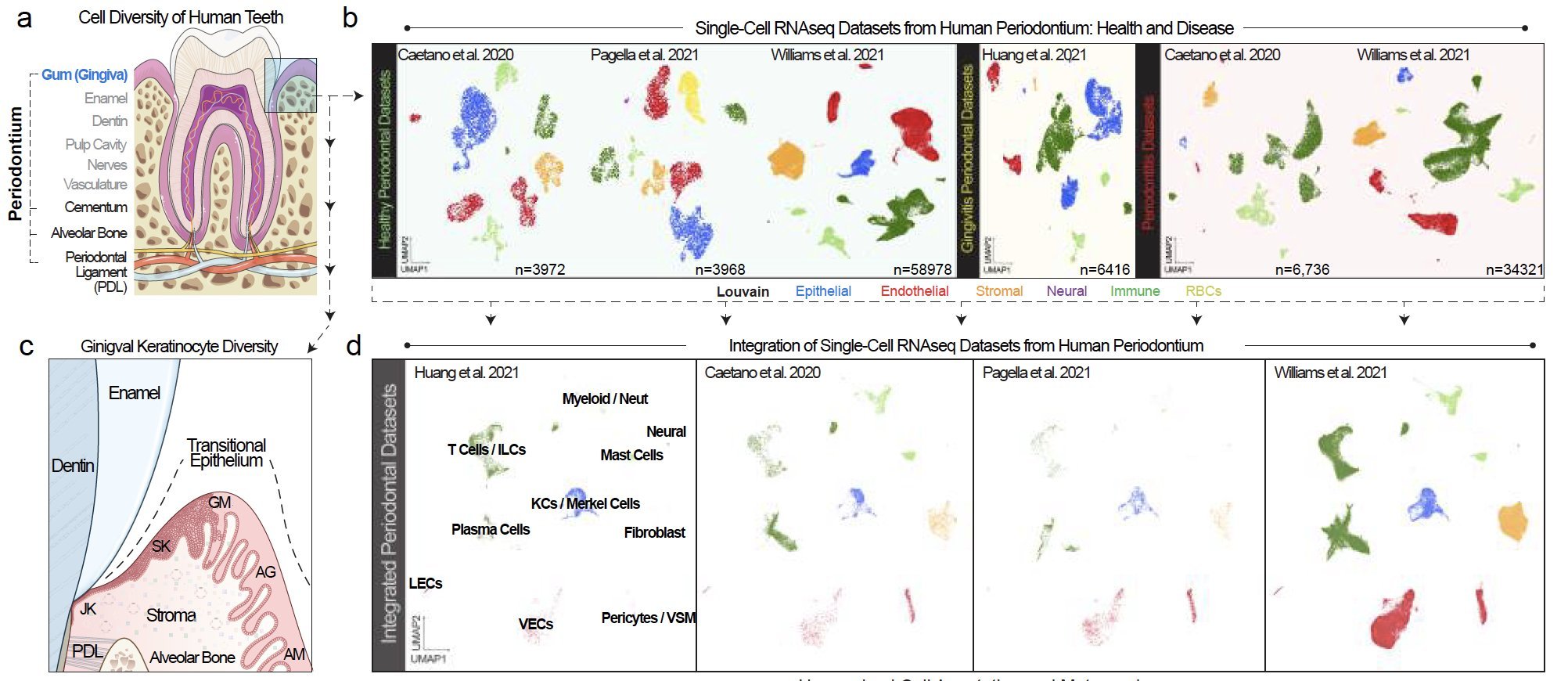
Scientist Kevin Matthew Byrd, DDS, PhD, founder of the Human Cell Atlas Oral & Craniofacial Bio-network, publishes pre-print article using CellenicsⓇ
In their pre-print article, entitled “Evidence for polybacterial intracellular coinfection of epithelial stem cells in periodontitis”, Quinn T. Easter and Kevin Matthew Byrd from the American Dental Association (ADA) Science & Research Institute and their team of collaborators present the first integrated periodontal meta-atlas of single-cell RNA sequencing data using CellenicsⓇ open source software.

From novice to expert: The ultimate single-cell RNA-seq analysis course
In the rapidly advancing field of biotechnology, single-cell RNA sequencing is pivotal for understanding cellular heterogeneity. However, many biologists find themselves daunted by the complexities of bioinformatics. Challenges range from coding hurdles to software selection and data interpretation. Biomage introduces a solution: “Mastering Single Cell RNA-seq Data Analysis With Cellenics®”. Tailored for biologists, this self-paced course offers over 10 hours of video lessons and comprehensive PDF materials, eliminating the need for coding expertise. The course covers everything from the basics of single cell RNA sequencing to data processing, exploration, and effective visualization. Start your journey in mastering single-cell RNA-seq data analysis by enrolling at courses.biomage.net.



The Human Cell Atlas and Beyond: An Introduction to Single-Cell Data Atlases
Single-cell atlases provide a comprehensive view of gene expression patterns in cells within a particular organ or tissue. These atlases usually are projects generated through collaborative efforts between multiple research groups and institutions, and can lead to ground-breaking discoveries. In this post, we wanted to highlight some of the key atlases containing scRNA-seq data!

Converting Parse Biosciences Evercode™ data to be compatible with Cellenics® analysis software
In this article, we explain how to convert the files from Parse Bioscience's Evercode™ platform into the 10x format files, which you can upload directly in Cellenics®.

Leveraging Public scRNA-seq Data: A Guide to Repositories and Resources
Welcome to the thrilling world of single-cell RNA sequencing (scRNA-seq)! This cutting-edge technology has revealed the intricate details of cellular heterogeneity and shedding new light on the mysteries of biology. From studies of cancer biology to investigations of neural networks, public scRNA-seq datasets cover a wide range of organisms, tissues, and conditions, providing a rich resource for researchers of all backgrounds. In this blog post, we'll take you through some of the key publicly available scRNA-seq resources, and show you how to start searching for single-cell public data!

Data security in the Biomage-hosted community instance of Cellenics®: frequently asked questions
We often get asked about data security in the community instance of Cellenics® that’s hosted by Biomage. We understand that data security is important to you. For example, you might have unpublished data or clinical data that is particularly sensitive. This article aims to answer some of the questions we’ve heard most frequently!

Cellenics® now supports data generated using BD Rhapsody™ technology
The BD Rhapsody™ Single-Cell Analysis System is able to perform high-throughput single-cell RNA sequencing , allowing researchers to analyze tens of thousands of cells in a single experiment. As of recently, Cellenics® users can upload files generated by the BD Rhapsody™ technology and perform scRNA-seq data analysis!

2022 in review - our top 5 blog posts
The beginning of the new year is always a good opportunity to take stock of the past year. One of the exciting things that happened in 2022 is the launch of our blog! It's been an exciting year for the field of single-cell research, and there is no doubt that it will continue to be a hot topic in the world of biological research! In 2022, we took time to highlight some of the topics and perspectives in this field.
With the start of the new year, we wanted to reflect on the most popular blog posts we have published so far!

Biomarker discovery using scRNA-seq: what's the big deal?
Biomarkers have the potential to save lives.
Biomarkers are simply measurable characteristics of organisms, so this may seem like an overstatement. However, the use of biomarkers is becoming increasingly important both in clinical research and practice.
Recently, there has been a growing interest in using single-cell technologies to identify novel biomarkers. Single-cell RNA sequencing (scRNA-seq) is a powerful tool for identifying gene transcripts within individual cells and can be used to detect changes in gene expression that may be indicative of disease or may predict progression or outcome.

scRNA-seq analysis with Cellenics®
In her article, Ziwen Cass Li describes her scRNA-seq analysis with Cellenics® step-by-step. She explores the question - what are the transcriptomic differences between the developing and adult mouse brains at the single-cell level?

Single-cell analysis overview by Prof. Peter Kharchenko
In his presentation, Prof. Peter Kharchenko gives an overview of single-cell transcriptomics analysis, a powerful method that has progressed rapidly in recent years. He discusses the growth of single-cell RNA sequencing (scRNA-seq), and the subsequent high data volume. He also considers different single-cell RNA-seq data processing tools, why they are needed, and what cons they have.

Iva Babukova - Can Open Source Cure Cancer | Turing Fest 2021
In her presentation at Turing Fest 2021, our co-founder and Chief Technology Officer Iva Babukova described how our team at Biomage shortened the time to analyze single-cell RNA sequencing data from several months to just a week. Read some of the highlights from her talk in this blog post!

Converting CSV/TSV files to upload to Cellenics®
In this article, we’ll show how to convert CSV/TSV files to the features/barcodes/matrix format to be able to upload and analyze your data using Cellenics.

Single-cell RNA sequencing data analysis: 6 tools you need to know
Have you ever wondered which single-cell RNA-sequencing (scRNA-seq) data analysis software to choose? We have compiled a list of the best scRNA-seq data analysis tools in 2024 to help you move your single-cell research forward.

How to demultiplex a Seurat object and convert it to 10X files for analysis in Cellenics®
A brief tutorial written in R to demultiplex a Seurat object and convert it to 10X files, which can be directly uploaded to Cellenics®.

Converting H5 files for analysis in the open source scRNA-seq data visualization tool Cellenics®
In this article, we’ll show how to convert H5 files to the features/barcodes/matrix format to be able to upload and analyze your data using Cellenics while we work on adding native support for H5 files.

How to demultiplex a count matrix and convert it to Cellenics®-compatible 10X files
A brief tutorial written in R to demultiplex an rds object and convert it to 10x files (count matrices), which can be directly uploaded to Cellenics®.
